Abstract
T-cell acute lymphoblastic leukemia (T-ALL) arises from the accumulation of genomic abnormalities and the malignant proliferation of immature T-cells. Despite recent advancements in understanding the genetic alterations driving T cell leukemogenesis, patients still suffer from recurrent relapses and treatment-related toxicities. Genome sequencing has revealed significant heterogeneity and important insights into the genetic landscape of T-ALL. Mutations in epigenetic modifiers are frequently observed and serve as an attractive target for novel therapeutic approaches.
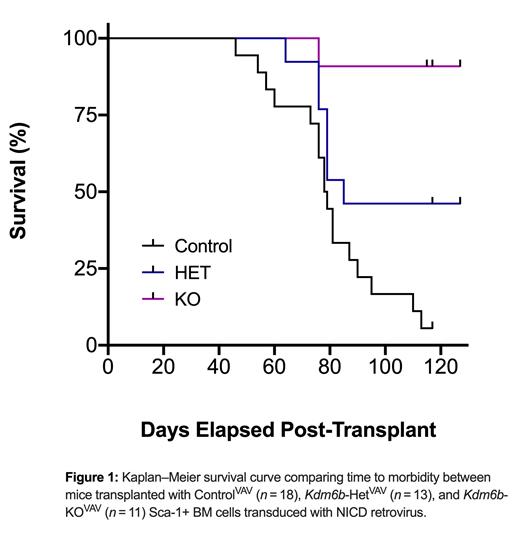
Histone demethylase enzymes play a critical role in the regulation of gene expression programs in T-ALL. KDM6A (UTX) is known to behave as a tumor suppressor in most T-ALL subtypes. However, it's gene paralog, KDM6B (JMJD3), is never mutated and can be significantly overexpressed in patients. Here, we show that KDM6B is required for T-ALL initiation. Using genetic mouse models, Sca-1 enriched WBM from Vav-Cre: Kdm6b+/+, Vav-Cre: Kdm6bfl/+, and Vav-Cre: Kdm6bfl/fl adult mice was transduced with a retrovirus expressing Notch1 Intracellular Domain (NICD). NOTCH1 gain-of-function mutations are the most frequent driver events in adult T-ALL, and this model recapitulates many of the human pathologies. Transduced cells were transplanted into irradiated mice. While there was robust engraftment in all groups at four weeks post-transplant, T-ALL cells were not sustained in the genetic absence of Kdm6b. Mice receiving control NICD-GFP+ cells succumbed to T-ALL with median survival of 79 days, whereas the only mice receiving Kdm6b-null NICD-GFP+ cells that developed disease were found to retain one copy of the Kdm6b floxed allele (Fig 1). To investigate the translational potential, we targeted KDM6B for genetic inactivation by CRISPR/Cas9 in primary T-ALL patient cells, followed by xenograft into NSG mice. The effect of KDM6B targeting was quantified over time by monitoring the variant allele fraction (VAF) of the transplanted cells. In most patient samples, KDM6B-targeted cells were significantly outcompeted over time, thus further highlighting the requirement of KDM6B in sustaining T-ALL tumorigenesis.
To examine the mechanism by which KDM6B sustain T-ALL cells, gene expression profiling was performed by RNA-seq on mouse T-ALL cells of genetic backgrounds Control, Kdm6b-Het, and Kdm6b-KO. Three gene sets were significantly downregulated in Kdm6b-deficient T-ALL cells compared to the Control group, all of which are involved in cell cycle processes. Additional validation of these findings with cell cycle functional studies is currently ongoing. Additionally, while Kdm6b has been described for its H3K27me3 histone demethylase function, recent studies have shown its involvement in regulating various gene expression programs through demethylase-independent mechanisms. To determine if the role of Kdm6b in T-ALL oncogenesis was related to it demethylase activity, we performed the same NICD retroviral transduction experiment with inclusion of a Kdm6b +/H1388Amouse genotype, a point mutation which renders Kdm6b catalytically dead. Our data shows that Kdm6b +/H1388Aphenocopies T-ALL with Kdm6b homozygous loss-of-function, showing no evidence of disease in the blood beyond 8-weeks post-transplant. We conclude from this that the leukemogenic role of Kdm6b requires it's H3K27me3 demethylase function. In summary, these data reveal Kdm6b as an oncogenic dependency in Notch1-driven T-ALL in both mouse and human systems, and present Kdm6b as a high value therapeutic target in adult T-ALL patients.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal